Google DeepMind reveals the shape of life
1 December 2020 | Written by Alberto Laratro
Google's artificial intelligence opens the door to a revolution in the biomedical field, determining the three-dimensional shape of proteins with unprecedented precision

“Medicine will change, research will change, bioengineering will change, everything will change”. The words of Andrei Lupas, evolutionary biologist of the Max Planck Institute reveal the extent of this achievement. The artificial intelligence developed by Google, DeepMind, through its AlphaFold program, has been able to solve one of the most important questions of modern biology: determining the three-dimensional shape of a protein from its amino acid sequence.
This is a challenge that has been going on for decades and that could have enormous repercussions in the medical field, and in any sector that concerns biology. The ability to predict the structure of a protein will allow accelerating, for example, the design of specific drugs, or to reveal the role of thousands of proteins within the human genome whose shape and function are still unknown today. opening the door to the understanding of those genes that can cause genetic diseases.
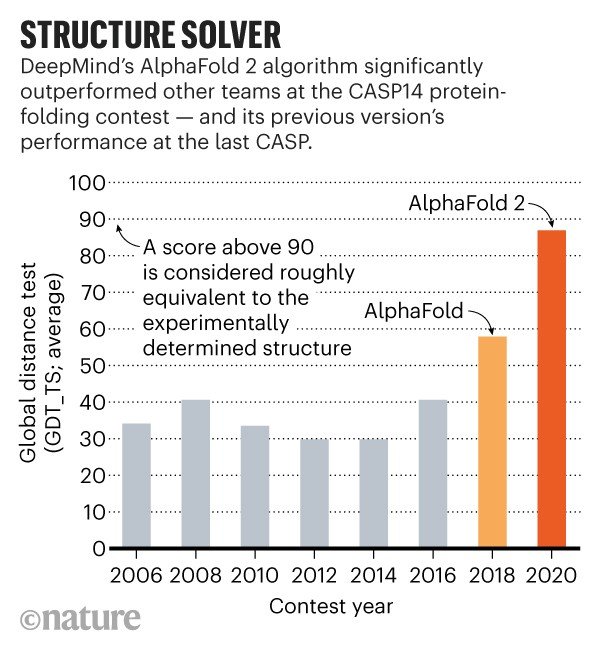
CASP, a competition to unveil the shape of life. The announcement of the results obtained by AlphaFold took place in the context of CASP, Critical Assessment of Structure Prediction, an international competition that since 1994 has seen research groups from all over the world compete to try to improve computational techniques to predict the structure of proteins. Google’s AI had already participated in 2018, placing itself well but without obtaining particular results. On November 30 this year, however, the announcement is moving research laboratories around the world. If on average the prediction methods and techniques are about 70% accurate, AlphaFold has reached 90%.
To date, X-ray crystallography is used to verify that the predicted structure corresponds to the real one, and AlphaFold will not replace these expensive and time-consuming methods but will provide researchers with new ways to approach research theoretically. before verifying the predictions experimentally, and will allow doing so at unparalleled speed.
The Lupas research team has worked for about ten years on the definition of the structure of a protein, also using crystallography, which, however, is not always able to reveal the form sought in a clear way. AlphaFold was able to rebuild it in half an hour.
The importance of proteins. Proteins are the building blocks of life, and their functions are determined by their shape. Guided by the atomic forces and by the “interlocking” between the different molecules, the amino acids, the proteins bend, deform and wrap themselves in a myriad of different shapes. Biologically speaking, life is a game of interlocking proteins.
Since the 1950s, when the first structure of a protein was defined thanks to X-ray crystallography, researchers have worked to understand how different combinations of amino acids can create such an enormous variety of shapes and therefore functions.
Developing a drug is a bit like creating a puzzle, but without knowing which pieces you have available to fill a certain empty piece. Accelerating the ability to predict the structure of a protein means accelerating the development of a drug, allowing us to understand more quickly which shape must have a piece to complete the puzzle.
Research becomes more and more “artificially intelligent”. The one used by the Google DeepMind team is not the only approach to making use of artificial intelligence. Half of the research teams that participated in this year’s CASP used Deep Learning, AlphaFold simply managed to do it more efficiently. Deep Learning, simplifying it, is the technique that allows you to train an artificial intelligence by providing it with a large number of examples to understand how a certain phenomenon works, in this case, the folding of proteins.
The use of these techniques is increasingly widespread and will play a fundamental role in biomedical research in the coming years. Demis Hassabis, co-founder and CEO of DeepMind said the company intends to make AlphaFold available and accessible. His previous version, in fact, was made public, allowing others to replicate his approach.
How laboratories will change. In early 2020 AlphaFold was able to predict the structure of some SARS-CoV-2 proteins before they were experimentally determined. Despite the competitive advantage of this technique, however, we will hardly find ourselves in front of a transformation in a short time of research laboratories: artificial intelligence techniques are already part of the tools used to study the structure of proteins. AlphaFold and other methods that exploit similar algorithms will complement and improve the techniques already in use, providing scientists with more and more methods